Beckmann lab
Molecular Infection Biology
IRI Life Sciences / HU Berlin
Molecular Infection Biology Group
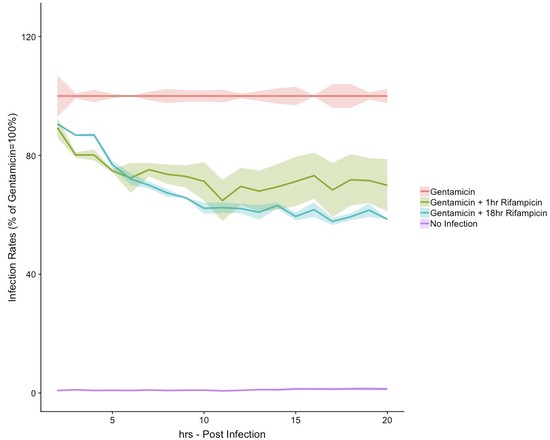
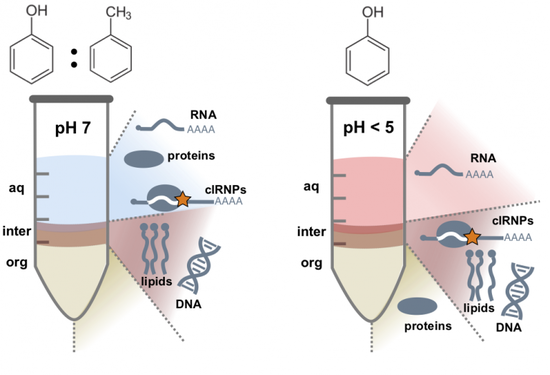
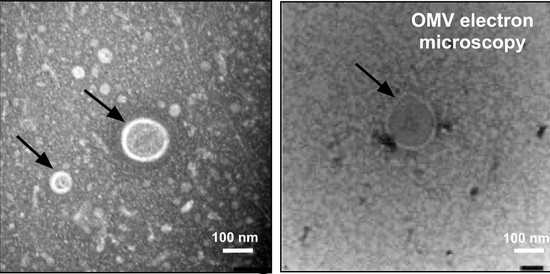
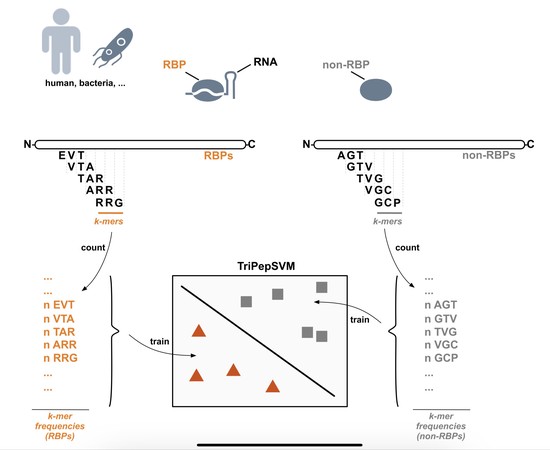
In our lab, we are investigating RNA-protein interactions. We recently developed a novel purification technique ( PTex ) which enables us to purify RNA-protein complexes from any biological source. We are using PTex to study post-transcriptional regulation of human cells which are infected with pathogenic bacteria. In our model organism Salmonella Typhimurium, we particularly look at extracellular vesicles and how the bacterium uses these to deliver RNA and proteins to host cells during infection.
When not at the bench, we play Pandemic Legacy. Or we do the Journal Club in a pub…